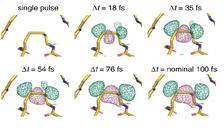
Difference density map of a thaumatin region with a bond between two sulphur atoms (dilsulfide bond). Negative (pink) and positive (green) structural differences, caused by stretching of the disulfide bond, increase with the delay between the two X-ray flashes. The other disulfide bonds in thaumatin show a similar effect. (Credit: Nass, Gorel, Abdullah et al., „Nature Communications“, DOI: 10.1038/s41467-020-15610-4, CC BY 4.0 [Source] )
X-ray lasers can reveal the spatial structures of biomolecules that are hard to come by with other methods. An international team of scientists also from DESY has now followed how the radiation damage - intense X-ray flashes vaporise these samples almost immediately - spreads in biomolecules to unprecedented detail. The results are important for the design and interpretation of X-ray laser measurements, as the team including DESY theorist, Beata Ziaja from the Center for Free-Electron Laser Science (CFEL) reports in the journal Nature Communications.
The spatial structure of proteins tells scientists how these biomolecules work and how they might be targeted by drugs. To resolve the structure, researchers X-ray small crystals grown from the protein under investigation. The crystals diffract X-rays in a characteristic way, and from the resulting diffraction pattern, the inner structure of the crystal and its constituents, the proteins, can be calculated.
Numerous protein structures have been solved this way, but some proteins grow only nanocrystals that need extra bright X-ray light to produce a usable diffraction pattern. X-ray free-electron lasers (XFELs) provide the brightest X-ray flashes on Earth and allow analysing even nanocrystals. Although the crystals are vaporised instantaneously, earlier research has shown that the diffraction pattern outruns the destruction, so the structure information can be retrieved before the crystal disintegrates. But just how the damage spreads in the proteins is largely unknown.
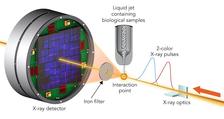
Thanks to a two-colour X-ray laser technique developed at the X-ray laser Linac Coherent Light Source LCLS at the US National Accelerator Laboratory SLAC in California, the team led by Ilme Schlichting of the Max Planck Institute for Medical Research and Sébastien Boutet of SLAC has now tested the technique to limits never before seen.
The scientists hit two types of crystallised biological molecules, the sweetener thaumatin and a complex of lysozyme (egg white) with the rare earth element, gadolinium, with pairs of X-ray laser pulses that had slightly different wavelengths and were up to 100 femtoseconds apart. A femtosecond is a millionths of a billionth of a second. The first pulse passed through the sample and was absorbed by a an iron foil. The second one had a slightly different colour that was not absorbed by the foil. It scattered off the sample, and entered a detector, forming a diffraction pattern that could be analysed to recreate the structure of the sample’s molecules and measure any changes caused by the first pulse.
“To obtain detailed microscopic insight into molecular processes induced by the first X-ray pulse, theoretical modeling was necessary,” explained Ziaja. The team found that the parts of a molecule that contain atoms heavier than oxygen absorbed the brunt of the X-ray damage. Chains of carbon atoms, which form the backbone of all proteins, also saw changes over time, but to a much smaller degree. These changes were not consistent throughout the entire molecule, occurring more in certain areas than in others, and they increased as the time between pulses was increased.
The scientists used two independent theoretical simulations: One from the DESY team, comprised of Ziaja, Malik Abdullah, Zoltan Jurek and Robin Santra, and one from the RMIT University in Melbourne and the University of Melbourne, to ensure the conclusions were not biased. “Both independent simulations confirmed the selectively progressing radiation damage which mostly affected sulphur ions, charging them strongly,” said Ziaja. “They also showed that the correlated dynamics of the highly charged sulphur ions were much slower than expected, due to the significant impact of the microscopic environment around the ions.”
These results show that in order to make reliable measurements, researchers need to model the specific parts of a sample rather than assuming all parts of the molecule are equally damaged. The study marks the beginning of a fuller understanding of how very short X-ray pulses produced by X-ray lasers modify the structure of biological molecules.
The team concluded that “diffraction before destruction” is an effective method of determining the structure of biological molecules, so long as scientists consider the intensity and duration of the pulses used to study them when interpreting their results. “Our analysis shows that a deeper understanding of the local damage effects is clearly needed to inform the design of XFEL studies of such systems and ensure the correct interpretation of the results to obtain accurate chemical information,” explained Ziaja.
Scientists from the Max Planck Institute for Medical Research, SLAC, RMIT University, the University of Melbourne, the Institute of Nuclear Physics of the Polish Academy of Sciences, the University of Hamburg and DESY contributed to this research.
(from DESY News)
Reference
Structural dynamics in proteins induced by and probed with X-ray free-electron laser pulses; Karol Nass, Alexander Gorel et al.; Nature Communications, 2020; DOI: 10.1038/s41467-020-15610-4