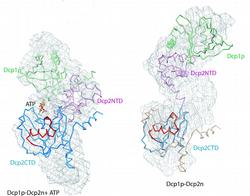
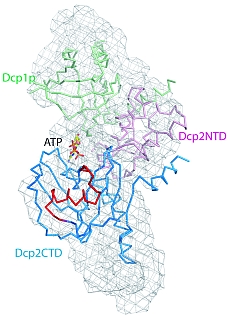
Figure 1: Structural models of Dcp1p-Dcp2n complex. The backbones of the crystal structures are superimposed onto the ab initio shapes generated from SAXS (shown as mesh). Left panel, closed crystallographic complex and SAXS model for the ATP-containing solution, right panel, open crystallographic complex and SAXS model for the ATP-free solution. Dcp1p is coloured green, the N- and C-termini of Dcp2n are displayed in pink (Dcp2CTN) and blue (Dcp2CTD), respectively, and the so-called “Nudix” motif is in red. In the left panel, ATP molecule is shown as sticks, and in the right panel, Dcp2CTD from the closed Dcp1p-Dcp2n complex is superimposed onto the open Dcp1p-Dcp2n backbone and coloured in wheat.
Meipei She1, Carolyn J. Decker2, Dmitri I. Svergun3, Adam Round3,4, Nan Chen1, Denise Muhlrad2, Roy Parker2 and Haiwei Song1,5
1 Laboratory of Macromolecular Structure, Institute of Molecular and Cell Biology, Agency for Science, Technology and Research, 61 Biopolis Drive, Proteos, 138673 Singapore
2 Department of Molecular and Cellular Biology, Howard Hughes Medical Institute, University of Arizona, Tucson, AZ 85721, USA
3 Hamburg Outstation, European Molecular Biology Laboratory, Hamburg 22603, Germany
4 Institute of Crystallography, Russian Academy of Sciences, Leninsky pr. 59, 117333 Moscow, Russia
5 Department of Biological Sciences, National University of Singapore, 14 Science Drive, 117543 Singapore
Published as: "Structural basis of Dcp2 recognition and activation by Dcp1", Mol. Cell 29, 337-349 (2008)
Small-angle X-ray scattering (SAXS) is rapidly becoming a streamline tool in structural molecular biology providing unique information about overall structure and conformational changes of native macromolecules in solution. Of particular interest is the joint use of SAXS with the high resolution methods like crystallography to study functional complexes. For a Dcp1p-Dcp2n protein complex playing a major role in the process of messenger RNA degradation, crystallographic analysis combined with SAXS and functional studies revealed that Dcp2p exists in open and closed conformations, the latter being the catalytically more active form. This suggests a decapping mechanism controlled by a conformational change between the open and closed complexes.
Tremendous progress has recently been achieved in structure determination of individual proteins due to large-scale initiatives in macromolecular X-ray crystallography (MX) [1]. Although high resolution information on isolated proteins is extremely valuable, it is also limited, as the most important cellular functions are accomplished by macromolecular complexes. The focus of modern structural biology is shifting towards the functional complexes [2], which are often of transient and flexible nature, hampering the growth of well-diffracting crystals. Even if the high resolution structure is solved, it represents a snapshot of the static structure in the crystal, which may be influenced by the crystallographic packing forces. Moreover, structural changes initiated by ligand binding or by alterations in external physical and chemical conditions are difficult to study in the crystal. Synchrotron small-angle X-ray scattering (SAXS) is a rapid technique to study overall structure of native macromolecules in solution. Recent progress in instrumentation and data analysis [3] significantly enhanced resolution and reliability of structural models provided by the technique. In the absence of other data, SAXS provides low resolution macromolecular shapes ab initio, but very important are its applications together with MX. Most notably, quaternary structure of complexes can be analysed by rigid body refinement using high resolution models of the subunits [3].
At the EMBL beamline X33 (DORIS III ring at DESY), SAXS is employed to validate and complement structural models provided by MX in numerous user projects. One of the examples is the study of a functional protein complex involved in messenger RNA (mRNA) degradation, an important cellular process controlling gene expression and the elimination of aberrant mRNAs [4]. A critical step in mRNA degradation is the removal of its 5‘ cap terminal structure, which is catalysed by a decapping complex consisting of the two proteins, Dcp1p and Dcp2p, where the latter is a catalytic subunit recognizing and cleaving the capped mRNA substrate. Their individual crystal structures have recently been solved [5,6].
Figure 2: Experimental and computed SAXS data. From top to bottom: Dcp1p-Dcp2n complex without and with ATP, and free Dcp2n without ATP (addition of ATP to Dcp2n yields the curve coinciding within the errors). The logarithm of the scattering intensity is plotted
against the momentum transfer s = 4π sinθ/λ, where 2θ is the scattering angle and λ = 1.5 Å is the X-ray wavelength. The plots are displaced along the ordinate for better visualization. Dots with error bars: experimental scattering; dashed lines: scattering from the models. Red: ab initio GASBOR models; green: open crystal complex, blue closed crystal complex (for Dcp2n, green and blue are extended Dcp2n as in the open crystal complex and compact free Dcp2n in the crystal, respectively).
A recent publication in February 2008 issue of Molecular Cell presents a joint MX, SAXS and biochemical study providing in sight into how Dcp1p interacts with and infl uences Dcp2p activity. The crystal structure of a complex of Schizosaccharomyces pombe Dcp1p with Dcp2n (a truncated version of Dcp2p) was solved at 2.8 Å resolution. Surprisingly, the asymmetric crystallographic unit contained two types of complexes, one with open, the other with closed conformation of Dcp2n (Figure 1). Moreover, an ATP molecule used as a crystallization additive was located in the catalytic centre of closed Dcp2n form but not in the open form. The closed conformation could correspond to the catalytically active enzyme, where the ATP molecule may be mimicking the interaction of the RNA substrate with Dcp2p. The observed effect could however also be an artifact generated by the crystal packing. Also, the open structure was incomplete with numerous missing loops in the C-terminus of Dcp2n.
SAXS was employed to study the solution structure of the Dcp1p-Dcp2n complex in the absence and presence of ATP (Figure 2). The radius of gyration Rg and the maximum size Dmax of the complex without ATP agree well with those of the open crystal form. Addition of ATP decreases the Rg by 5 Å and Dmax by about 10 Å making the parameters compatible with those of the closed crystal form. The low resolution shapes of the complex with and without ATP reconstructed ab initio from the scattering patterns agree well with the closed and open crystal structures, respectively (Figure 1). Moreover, the experimental data in the presence of ATP is neatly matched by the scattering pattern computed from the closed model but not from the open model (Figure 2). In contrast, the measured scattering from the ATP-free complex agrees better with the pattern computed from the open form (after addition of the missing loops using the coordinates of the Dcp2n C-terminus in the closed form) but displays some deviations suggesting that the structure in solution may be flexible.
For the Dcp2n alone (Figure 2), addition of ATP did not prompt a change in the SAXS scattering pattern. The overall shape of free Dcp2n agrees well with the extended Dcp2n conformation as in the open crystallographic model of the complex, but not with the compact structure of free Dcp2n in the crystal [6]. This indicates that Dcp2n exists in an extended conformation in solution and also that the closure of Dcp2n requires its complex with Dcp1p.
These structural results were further corroborated by biochemical assays and mutagenesis demonstrating the higher catalytic activity of the close form. One may therefore conclude that Dcp1p activates Dcp2p by stabilizing a closed conformation of the complex and that a conformational change between these open and closed complexes might control the mRNA decapping mechanism. Overall, the study underlines the power of the joint use of MX and SAXS in the study of functional macromolecular complexes.
|
References |
||||||||||||
|
||||||||||||
|
Contact information |
||||||||||||
|
Dmitri Svergun |
||||||||||||
| Further Information |