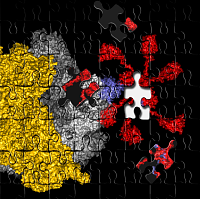
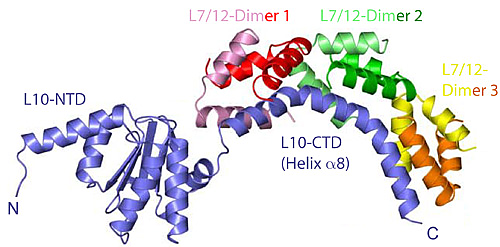
Figure 1: The structure of the heptameric complex of L10 (blue) and the 6 copies of the L7/12 N-terminal domain.
Frank Schlünzen1,2, Jörg M. Harms1,2, Markus Wahl3
1Max-Planck-Institute for Molecular Genetics, Berlin
2MPG-ASMB, c/o DESY, Hamburg
3MPI for Biophysical Chemistry, Göttingen
Published as: M. Diaconu et al., Cell 121, 991-1004 (2005)
Genomes of rather divers species like humans, mice or worms have been found to be amazingly similar. In these and all other organisms, the genetic information is translated by the ribosome, in a way that the small but crucial differences are accurately maintained. To understand how the ribosome works, and how antibiotics block specifically bacterial ribosomes, molecular structures of the ribosome and the related co-factors are essential. In a collaborative study employing different physical and biochemical methods we were able to add another piece to the ribosomal puzzle, by a detailed characterization of one of the key elements of the ribosome, the so called L7/12 lateral stalk of the ribosome.
The ribosome is a huge molecular machine, composed of 3 strands of ribosomal RNA and roughly 50 different proteins or altogether about 200.000 atoms. The most important reactions catalyzed by the ribosome are however extremely well localized in the three main functional centers of the ribosome: the decoding center (DC) on the small or 30S subunit, responsible for the correct translation of the genetic code; and the peptidyl transferase center (PTC) of the large or 50S subunit, gluing the individual amino acids into a polypeptide chain. Both these functional centers have been well characterized in a series of crystallographic studies on the 30S and 50S subunits as well as the full 70S ribosome (see 1, 2 for recent reviews). The third functional center is the GTPase associated region (GAR), stimulating GTP hydrolysis of elongation and initiation factors. Without the concerted action of the initiation and elongation factors, the ribosome would be a deadly slow and faulty molecular machine.
The GAR is comprised of distinct structural elements of 23S rRNA and ribosomal proteins L10, L11, L7/12 forming the lateral ribosomal stalk. All of these elements are virtually invisible in any of the available ribosome structures, and the underlying mechanisms and interactions consequently escaped up to now a detailed characterization. The reason for the lack of structural data was illustrated by a number of electron microscopy and NMR studies, which demonstrated the puzzling flexibility of the lateral stalk (reviewed in 3, 6). This inherent flexibility made it very difficult to visualize the structure of the stalk with protein crystallography or electron microscopy.
Recently, however, Markus Wahl and co-workers (MPI for Biophysical Chemistry, Göttingen) determined the crystal structure of protein L10 in complex with the N-terminal domains of L7/12 (Figure 1) using x-ray data collected at BW6/HASYLAB to a resolution of 1.9Å (4). Unexpectedly, the structure from the thermophilic bacterium Thermotoga maritima contained 6 copies of L7/12 although it was generally believed that ribosomes from all organisms contain exactly 4 copies, regardless of the species. However, it turns out that solely the length of the C-terminal α-helix of L10 determines the number of copies. The unexpected variation in the number of L7/12 copies bound to L10 was confirmed by antibody labeling and independently with mass-spectroscopy (5). Increased L7/12 copy number may somehow correlate with the adaption of an organism to its specific environment. Thus, it is possible that the additional copies of L7/12 facilitate the co-factor recruitment under harsh conditions like elevated growth temperatures.
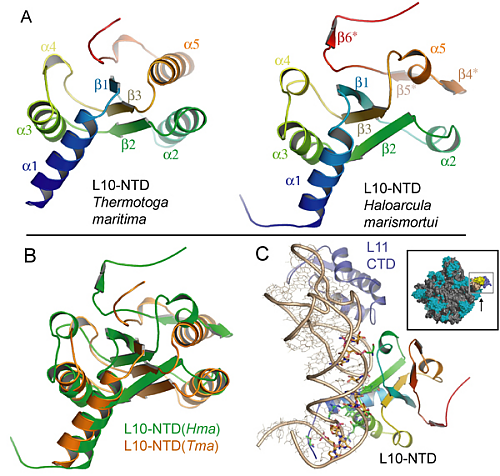
However, the structure was solved off the ribosome, but to shed light on the GTPase mechanisms, the structure had to be anchored onto the ribosome. Fortunately, we were also able to deduce the structure of the N-terminal part of L10 in the 50S ribosomal subunit from Haloarcula marismortui (Figure 2), which became possible by applying generalized 3-D image processing filters. The results obtained for the archaeal L10 turned out to be surprisingly similar to the eubacterial L10, despite the marginal sequence homology between the two proteins (Figure 2).
This agreement nicely served as a cross-validation but also explains the long-known similarity in the organization of the lateral stalk from very different organisms. The high structural homology made it simple to dock the complete L10/L12 stalk onto the ribosome, and the interpretation agreed well with electron microscopy images of the 70S-EF-G complex as shown by Holger Stark and co-workers. To elucidate the interaction of L7/12 with elongation factor EF-G a number of mutational and kinetic experiments have been performed by Marina Rodnina’s group, which confirmed that the dissociation of L7/12 from the ribosome, as observed under certain growth conditions, does not contribute to co-factor recruitment. This means, that L7/12 does not hunt for co-factors in the cytoplasmic space but rather remains anchored to the ribosome and fishes for co-factors using the multiple, flexible L7/12 hooks, which drastically accelerates co-factor association.
Methods
X-ray data were exclusively collected at the MPG-beamline BW6 at DORIS/HASYLAB. The structure of the L10-L7/12 complex was solved by using SeMet-MAD phasing (4). The determination of the structure of L10 within the Haloarcula marismortui 50S ribosomal subunit determined by us, was based on the published structure (reviewed in 7) using 3D images processing filters for improving the electron density maps. EM reconstructions of the 70S-EF-G complex were performed in the group of Holger Stark, MPI for Biophysical Chemistry in Göttingen, and the biochemical and kinetic analysis was done by Marina Rodnina and co-workers, Institute of Physical Biochemistry, University of Witten/Herdecke.
| References |
|
[1] Liljas, A. (2004) Structural aspects of protein synthesis. World Scientific Publishing Co. Pte. Ltd., Hackensack, NJ 07601, USA; pp. 290. |
| Contact information |
|
F. Schlünzen |
| Further Information |