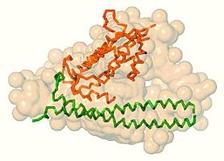
Figure 1: Solution scattering results for the longer complex Cdt1:Gem. Black dots denote experimental SAXS data, the green and the red lines are the fits from the heterotrimer and heterohexamer, respectively. Ab initio shape is shown as yellow beads and the crystal structure of the hexamer as a backbone, whereby two Geminin dimers and two copies of Cdt1 are highlighted by different colours.
V. De Marco1, P. J. Gillespie2, A. Li2, N. Karantzelis3, E. Christodoulou1, R. Klompmaker4, S. van Gerwen1, A. Fish1, M. V. Petoukhov5, M. S. Iliou6, Z. Lygerou6, R. H. Medema4, J. J. Blow2, D. I. Svergun5, S. Taraviras3, A. Perrakis1
1. Department of Biochemistry, Netherlands Cancer Institute, Plesmanlaan 121, 1066CX Amsterdam, The Netherlands
2. Wellcome Trust Centre for Gene Regulation and Expression, College of Life Sciences, University of Dundee, Dundee DD1 5EH, United Kingdom
3. Department of Pharmacology, Medical School, University of Patras, 26500 Rio, Patras, Greece
4. Department of Medical Oncology and Cancer Genomics Center, Laboratory of Experimental Oncology, University Medical Center Utrecht, Universiteitsweg 100, 3584 CG Utrecht, The Netherlands
5. European Molecular Biology Laboratory, Hamburg Outstation, Notkestrasse 85, D-22603 Hamburg, Germany
6. Department of Biology, Medical School, University of Patras, 26500 Rio, Patras, Greece
Published as: “Quaternary structure of the human Cdt1-Geminin complex regulates DNA replication licensing”, Proc. Natl. Acad. Sci. USA 106, 19807 (2009).
Small angle scattering becomes a streamline tool in modern structural molecular biology providing valuable information about overall structure and conformational changes of native individual proteins and functional complexes. The variety of questions addressed by the technique ranges from evaluation of the overall geometrical parameters and low-resolution shape reconstruction to structure validation and molecular modelling. Recently the method was applied in combination with X-ray crystallography for a structural study of the human Cdt1-Geminin complex regulating DNA replication licensing which is essential for genomic integrity. It was found that the transition of the complex from heterohexamer to heterotrimer acts as a switch between inhibitory and permissive states for DNA replication.
Recent progress in instrumentation and automation of the X33 SAXS beamline [1], together with the development of very efficient methods for solution scattering data analysis [2], attracts steadily growing interest from the biological user community in Europe and overseas. Solution scattering profiles are rapidly collected from biological macromolecules and multicomponent assemblies at native conditions and interpreted using efficient algorithms [3]. The modern computational tools allow determination of particle shape at low resolution in the absence of a priori structural information, discrimination between concurrent three-dimensional atomic models and rigid body refinement, as well as quantitative analysis of equilibrium mixtures and flexible systems. Complementary information from other methods (bioinformatics, electron microscopy, NMR, biochemistry, crystallography, neutron scattering) is readily incorporated into the modelling procedures [4], and this significantly improves the reliability of the structural models. A synergistic approach employing SAXS, crystallography and functional studies was recently applied in collaboration with the group of A. Perrakis from the Netherlands Cancer Institute (NKI) to shed a light on DNA replication licensing.
The duplication of chromosomal DNA is an essential process for all organisms, which has to be tightly regulated. Origin licensing in eukaryotic cells prohibits unwanted replication of DNA segments in a single cell cycle. A protein called Cdt1 plays a key role in control and regulation of this process in space and time. The tight spatio-temporal control of Cdt1 is partly achieved by binding of its inhibitor, Geminin (Gem). In this study, the molecular shapes of two Cdt1:Gem complexes, including Geminin truncated in two different lengths, were analysed.
The overall structural parameters obtained from the recorded SAXS profiles of the longer complex agree well with a heterohexameric assembly formed by two Cdt1 molecules and four copies of Geminin (2x[Cdt1:2xGeminin]). The crystallographic model of the heterohexamer determined by the NKI group consists of two heterotrimers related by a two-fold crystallographic symmetry axis. The scattering curve computed from such a hexamer yields a good fit to the experimental scattering data and was crucial to confirm that this crystallographic packing is also observed in solution. The model of the crystallographic hexamer is compatible with the low resolution shape reconstructed ab initio from SAXS (Fig. 1). At the same time the solution scattering experiment suggested that in the case of the truncated complex, the heterohexamer is not formed. Instead, the overall size and mass evaluated from the SAXS data point to a lower molecular weight assembly, a heterotrimer (Cdt1:2xGeminin). Indeed, the heterotrimer extracted from the crystal structure of the heterohexamer (with truncation of the C-terminal portion of Geminin) can be accommodated very well within the SAXS-based shape and it also provides a good fit to the experimental profile (Fig. 2). The fits computed from the heterotrimer model to the longer complex data (Fig. 1) and from the heterohexamer to the truncated complex data (Fig. 2) are rather poor, confirming the discriminative power of SAXS to detect different oligomeric states.
The above findings show that the presence of the C-terminal residues of Geminin coiled coil is critical for the quaternary assembly of the native complex. At the same time, it was shown by functional studies that the overall organisation of the complex is important for the inhibition of Cdt1 activity. Geminin capable of forming the heterohexamer was able to inhibit DNA licensing, while the shorter Geminin construct unable to support heterohexamer formation showed diminished inhibitory activity. This allowed us to conclude that the Cdt1:Gemin complex can exist in a heterotrimeric, “permissive”, state that allows DNA replication, and in heterohexameric, “inhibitory”, state that would not allow the formation of the full DNA replication license. This result helps to understand how the duplication of chromosomal DNA is regulated during the cell cycle.
| References | ||||||||
|
| Contact information |
|
Dmitri I. Svergun |
| Further Information |