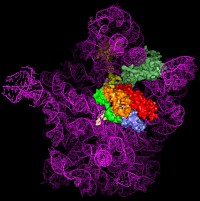
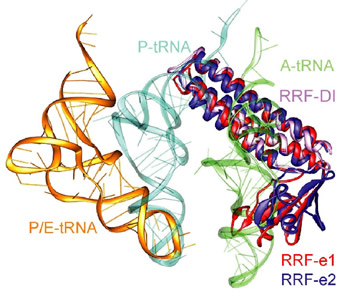
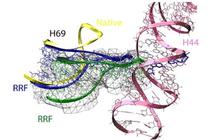
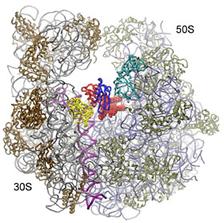
Figure 1: Comparison of different tRNA binding sites with the possible conformations of RRF on the ribosome. RRF overlaps with both A- and P-site tRNA binding sites. Binding of RRF is therefore concurrent or subsequent to translocation of P-site tRNA to the P/E-hybrid state.
Daniel N. Wilson1, Frank Schlünzen1,4, Jörg M. Harms2, Takuya Yoshida3, Tadayasu Ohkubo3, Renate Albrecht1, Jörg Buerger1, Yuji Kobayashi3, Paola Fucini1
1Max-Planck-Institute for Molecular Genetics, Ihnestr. 73, D14195 Berlin, Germany
2Riboworld.com, Seestrasse 8b, 22607 Hamburg, Germany
3Graduate School of Pharmaceutical Sciences, Osaka University, 1-6 Yamadaoka, Osaka, Japan.
4MPG-ASMB, c/o DESY,Notkestr. 85, 22603 Hamburg, Germany
Published in: Embo J. 24(2) 251-260, 2005
The ribosome deciphers and translates the genetic code in all living organisms. This process - termed protein biosynthesis – requires several co-factors accelerating or catalyzing various steps. Termination is the last step of protein biosynthesis and requires the combined action of the Ribosome Recycling Factor (RRF) and the Elongation Factor G (EF-G). We recently determined the crystal structure of RRF bound to Deinococcus radiodurans 50S subunit at 3.3 Å resolution. These results provide a high-resolution structural description of the specific interaction of RRF with the ribosome and the conformational changes induced by RRF. The data permit to derive a accurate model for the synergistic action of EF-G and RRF during termination of protein biosynthesis and ribosome recycling.
The ribosome deciphers and translates the genetic code in all organisms. Ribosome recycling is the final stage of translation and involves the concerted action of the ribosome recycling factor (RRF) and elongation factor G (EF-G) to disassemble the post-termination complex for the next round of translation. Although RRF was discovered in the early 1970s [1], the exact mechanism by which RRF mediates ribosome recycling still remains to be fully elucidated.
RRF is universally conserved in bacteria, but not present in archaea or eukaryotes (with the exception of chloroplast and mitochondrial RRFs). Deletion of frr, the gene encoding RRF, is lethal to E. coli cells [2] and, in the absence of RRF, ribosomes remain bound to the mRNA and initiate spontaneous translation downstream of the stop codon [3]. The cellular importance and kingdom distribution of RRF make bacterial ribosome recycling an attractive target for drug design; however, such an undertaking requires an atomic understanding of the ribosomal binding site of RRF.
Despite having a shape remarkably similar to tRNA [4], recent studies using hydroxyl radical probing and cryo-EM have revealed that RRF adopts a significantly different orientation on the ribosome (Figure 1) to that of a tRNA [5,6]. In this orientation domain I of RRF interacts predominantly with the large subunit, rather than with the small subunit as predicted by tRNA mimicry, which is in agreement with the observation that domain I alone (RRF-DI) can bind to the 50S subunit with an affinity comparable to that of RRF. Though biochemical methods and cryo-EM provide a good model for the location of RRF on the ribosome, the precise mechanism of termination remained open due to the rather limited spatial resolution of these methods.
We have recently determined the crystal structure of domain I of E. coli RRF bound to the D. radiodurans 50S ribosomal subunit at 3.3 Å. RRF-DI is located on the intersubunit side of the 50S subunit, in a position very similar to the positions derived from the cryo-EM and hydroxyl radical probing studies. However small differences in orientation are observed, which have some important consequences with respect to the interaction of specific residues of RRF with the amino acid of the ribosomal proteins and nucleotides of the 23S rRNA.
A detailed characterization of the interactions between RRF and the large subunit provides support for the translocation of RRF through the ribosomal particle, despite its different positioning to tRNA: All the contacts observed between RRF and the large subunit encompass elements involved with either the correct positioning of the tRNA on the ribosome or translocation of tRNA through the ribosome, such as ribosomal proteins L16, L27 and rRNA helices H80, H93 and H69-H71. The most extensive interaction that RRF has with the large subunit are with H69 and H71, which are the large subunit components of intersubunit bridges, that are involved in translocation of tRNA. The intersubunit bridges connect the 30S and 50S ribosomal subunits forming the (active) 70S ribosomes. The integrity and conformation of the bridges are therefore crucial for the stability of the 70S ribosome.
Interestingly, in our 50S-RRF complex H69 has also undergone a substantial conformational change, resulting in the repositioning of the tip of H69 and movement by 20 Å toward the small ribosomal subunit (Figure 2). In this position on a 70S ribosome, the tip of H69 would clash with its counter-bridge element h44 of the small subunit. The observed movement and novel fold of H69 induced by RRF binding to the 50S subunit presumably represent the same conformational changes that RRF would try to induce in a 70S ribosome. Consistently, rearrangement within this region was also observed by the cryo-EM study of RRF-70S complex [5].
Because RRF requires the action of EF-G to complete its function, we fitted the crystal structure of EF-G on the RRF-50S structure according to the position observed in the 70S•EF-G•GDPCP complex [7]. Clashes between domain II of RRF and domain III and IV of EF-G suggest that upon EF-G binding, domain II of RRF is pushed further toward h44 in the 30S subunit. Thus, the combined action of EF-G and RRF on h44-H69 may weaken the intersubunit bridge leading to subunit dissociation under some circumstances (Figure 3).
METHODS
E. coli RRF-DI was prepared as described previously [8]. D. radiodurans 50S subunit crystals [9] were soaked in a solution containing 5-10 µM RRF-DI for 24 hrs, prior to freezing. The synchrotron experiments were carried out at beam line facilities at the ESRF, Grenoble/France, the SLS, Villigen/Switzerland and at BW6/Hasylab/DESY, Hamburg/Germany.
| References |
|
[1] Hirashima, A., and Kaji, A. Biochem. Biophys. Res. Com., 1970, 41, 877-883. |
| Contact information |
|
F. Schlünzen |
| Further Information |