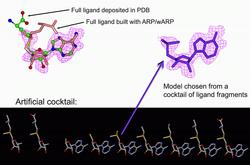
Figure 1: Building partially ordered ligands. Top left: the difference density map in the area of the ligand; Adenosylmethionine as in the PDB model (1v2x); full ligand built with ARP/wARP – none of the two models fit the density well. Bottom: an artificial cocktail of ligand fragments. Top right: a partial ligand built with ARP/wARP, matching the density.
Gerrit G. Langer1, Guillaume X. Evrard1, Venkataraman Parthasarathy1 and Victor S. Lamzin1, Serge X. Cohen2, Krista Joosten2 and Anastassis Perrakis2
1European Molecular Biology Laboratory, c/o DESY, Notkestrasse 85, Hamburg 22607, Germany
2Department of Molecular Carcinogenesis, Netherlands Cancer Institute, Plesmanlaan 121, Amsterdam 1066CX, the Netherlands
Published as: “Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7”, Nature Protocols 3, 1171-1179. PMID: 18600222 (2008).
ARP/wARP is a software suite to build macromolecular models in X-ray crystallographic electron density maps. Structural genomics initiatives and the study of complex macromolecular assemblies and membrane proteins rely on advanced methods for 3D structure determination. ARP/wARP 7.0 meets these needs by providing the automated tools for: iterative protein model building including a high-level decision-making control module; fast construction of the secondary structure of a protein; building flexible loops in alternate conformations; placement of ligands, including a choice of the best-fitting ligand from a ‘cocktail’; and finding ordered water molecules. All protocols are easy to handle by a non-expert user and the time required is typically a few minutes, although iterative model building may take a few hours. ARP/wARP is a continuous collaborative project between two groups at the EMBL in Hamburg and the NKI in Amsterdam.
Structural genomics initiatives and medically oriented highthroughput structure determination projects emphasise the need for advanced methods for structure determination [1]. In X-ray macromolecular crystallography, availability of comprehensive software packages has had a major impact on structural biology research. Crystallographic model building has been traditionally done by expert users, with the aid of specialised interactive graphics software. The automation of this process and linking model building and refinement together into a unified process, first exemplified in the ARP/wARP package [2], was followed promptly by significant developments worldwide. ARP/wARP has been used extensively for thousands of structure determinations. Examples include a three-protein complex that is crucial in chromosome segregation (complete list of publications); complexes in spindle assembly checkpoint formation; ubiquitin conjugation; transcriptional regulation of mRNA; cargo transport along micro tubules; studies of bioluminescence; the structural dissection of an enzyme involved in the synthesis of inflammatory mediators; ligand recognition by lipoprotein receptors; investigation of membrane-binding proteins in signal transduction in photo-response, the plant aquaporin mechanism and the functional characterisation of a prokaryotic Ca2+-gated K+ channel. Moreover, ARP/wARP is often used as a benchmark to evaluate the quality of electron density produced by new methods [3] and has been integrated in many crystallographic software pipelines as the default model-building engine [4]. Successful automated building of a considerable part of models is now possible at a resolution as low as 2.7 Å [5] and a wide spectrum of ARP/wARP functionalities is outlined below.
Figure 2: Remote execution of ARP/wARP protein model building directly through the web link:
http://cluster.embl-hamburg.de/ARPwARP/remote-http.html.
Free atoms and hybrid models. Firstly ARP/wARP condenses the map information to a set of identical free atoms that represent the electron density. As model building iteratively proceeds, some free atoms are recognised as part of a protein chain and gain chemical identity. This mixture is an ARP/wARP hybrid model that incorporates chemical knowledge from the partially built protein model, whereas its free atoms continue to interpret the electron density in areas where no model is yet available.
Main chain tracing in ARP/wARP uses all atoms of the hybrid model as potential Cα atoms. Peptide units are recognised by matching the surrounding electron density to that precomputed from known structures. The recognised peptides are assembled into linear chain fragments with partial ‘guessed’ side chains using a limited depth-first graph-search algorithm.
Side chains are subsequently docked in protein sequence and built in the best rotamer configuration.
Loop building. Using the sequence docking and a distribution of penta-peptide fragments derived from known structures, several loop conformations are constructed and the one fitting best the density is chosen. This helps model building in low density areas.
Secondary structure recognition. At a resolution where electron density maps lack atomic features sparse map grid points with 1 Å spacing are selected as potential Cα atoms. After successive filtering steps trace assemblies of helical or stranded fragments are generated and averaged. Peptide backbone and Cβ atoms are added and the most likely chain direction is selected. The procedure is applicable to a resolution as low as 4.5 Å.
Ligand building. When the protein structure is completed, bound ligands or cofactors can be built in the difference electron density map. First, regions of density that have the same volume as the ligand are identified. Then we use numeric features of the density region and its sparse representation to produce an ensemble of putative ligand structures. The single best model is chosen after refinement of the whole ensemble [6].
Cocktail screening. The comparison of the shapes of electron density blobs with the shape of the search ligand is used to distinguish compounds from a list (cocktail) of candidates. The ligand that fits best is selected for its further construction. An application of the cocktail screening for building a partially ordered ligand is exemplified in Figure 1.
Solvent building. After the protein part of the model is complete, a solvent structure can be constructed in the electron density map.
Availability and use of the software. ARP/wARP is freely available to all users and free of charge for academic usage, http://www.arp-warp.org. Software installation is user friendly and a User Guide can be referred to for additional information. There are no software dependencies other than the CCP4 package and particularly the program REFMAC6. ARP/wARP model building can be used after experimental phasing or molecular replacement. ARP/wARP can run through the CCP4i interface or be launched from the command line. A protein modelbuilding task can be submitted to a 64-processor Linux cluster at the EMBL Hamburg; the results can be viewed through a web browser, http://cluster.embl-hamburg.de/ARPwARP/remote-http.html, Figure 2. Typically, the secondary structure tracing and ligand building run within a few minutes. Model building for a 500-residue protein takes about an hour and the time needed scales about linear with the size of the structure.
|
References |
||||||||||||
|
||||||||||||
|
Contact information |
||||||||||||
|
Victor Lamzin |
||||||||||||
| Further Information |