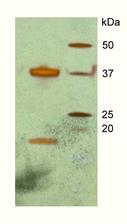
Figure 1: A silver-stain gel from crystals of the DAP kinase–CaM complex, indicating the presence of calmodulin (arrow) in the crystals in addition to DAP kinase.
Inaki de Diego1, Jochen Kuper2, Neda Bakalova1, Petri Kursula3, Matthias Wilmanns1
1. EMBL-Hamburg c/o DESY, Notkestrasse 85, D-22603 Hamburg, Germany
2. Rudolf Virchow Center for Biomedical Research, University of Würzburg, Versbacher Str. 9, D-97078 Würzburg, Germany
3. Department of Biochemistry, University of Oulu, P.O. Box 3000, FIN-90014, Finland
Published as: “Molecular basis of the death-associated protein kinase-calcium/calmodulin regulator complex”. Sci. Signal. 3, ra6 (2010).
Investigating how cells communicate is fundamental to our understanding of biology. We have determined the first molecular structure of a protein kinase in complex with the universal regulator protein calmodulin and calcium. The structure, which provides a model for many other protein kinases, allows insight into how its function could be specifically blocked. Since the selected protein kinase is associated with a number of frequently occurring tumours, our data provide a basis for developing new anti-cancer therapies.
Signalling processes in cells are essential for their development and for their function in the mature state. Alterations of signalling often lead to degenerate functional outcomes, which in frequent cases cause cancer. The knowledge of the underlying molecular mechanisms is essential to develop new therapies against these malignancies. To gain insight into one of these signalling systems, we have been studying a model system belonging to the so-called calcium / calmodulin (CaM) dependent protein serine/threonine kinases. Kinases, which make up 2% of all proteins from the human genome, are enzymes that catalyze phosphorylation reactions. Collectively they form the “kinome”. Phosphorylation is done by transferring a phosphate group from an adenosine-5’-triphosphate (ATP) molecule, known as the “energy currency” of living systems, to a target protein. The fast and reversible nature of phosphorylation, together with the organization of kinases into hierarchical pathways or cascades, results in the high sensitivity and strong amplification of the input signals. As a consequence, of this kinases are of great importance in cellular pathways, especially signalling. With more than 70 predicted members, the CaM-dependent kinases form one of the largest superfamilies of the complete kinome. Inside the cell these kinases rely on intracellular calcium levels. This occurs through the modulator protein calmodulin, a universal intracellular calcium receptor, which picks up this signal and binds with the appropriate kinase. This complex can then become an active player in the cell’s machinery.
We chose a specific member of the CaM-dependent kinase family called the Death Associated Protein (DAP) kinase to study how calmodulin binds and regulates these kinases. DAP kinases have a central role in cell death pathways, including apoptosis, autophagy and necrosis. It has also become a key marker in cancer screens due to its tumour suppressor activity. It sup-presses tumour growth by inhibiting cell adhesion and migration and promoting cell death. Mutated versions of DAPK are known to be involved in the development of some cancers such as chronic lymphocytic leukaemia – one of the most common types of adult leukaemia and currently incurable. In contrast, in certain types of prostate cancer the role of DAP kinase seems to promote tumour growth when expressed at high levels. Knowing more about DAP kinase and how it functions may bring us a step closer to new cancer treatments. Since DAP kinase has physical similarities to other kinases controlled by calmodulin, we hope that this can also be used as a model for other similar systems.
In recent work, we first needed to establish protocols to express and purify DAP kinase / CaM as a stable complex that is maintained even under harsh crystallisation conditions. This is a prerequisite for high-resolution structural analysis by X-ray crystallography. We succeeded to crystallize a fragment of DAP kinase-1, which covers the catalytic kinase domain and a neighbouring domain that is responsible for CaM binding, together with CaM (Figure 1). As in so many other examples, the crystal structure, which we determined with X-ray data from the EMBL beamlines at DORIS III and from the ESRF, offered unexpected surprises (Figure 2). Although previous biochemical and cell biology data had indicated that CaM binding would lead to activation of the kinase, in the 3D structure of the complex we observed that CaM sits just in front of the active site of DAP kinase, thus blocking access to the kinase’ substrates - ATP and the protein target for phosphorylation. Therefore, our structure – although an exciting breakthrough as the first ever assembly of a protein kinase-CaM complex – immediately triggered new questions which we have started to solve more recently.
We are now employing a number of complementary structural biology techniques, such as fluorescence resonance energy transfer (FRET) spectroscopy and small angle X-ray scattering (SAXS) to explore the dynamic conformational space of this kinase, which leads either to its activation or deactivation. We are planning to exploit our knowledge to identify small molecule compounds that will specifically inhibit key steps in this activation / deactivation process. Embedded in several collaborations with colleagues from Israel, Norway and Germany, we are ultimately planning to make our knowledge useful for the development of new therapies in the clinic.
| Contact information |
|
Matthias Wilmanns |
| Further Information |