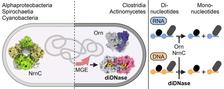
DiDNase (from the figure below). (Credit: CSSB)
Researchers at DESY Photon Science, together with international colleagues, have discovered a new class of enzymes that can be used to uncover previously unknown biological processes. The researchers suspect that these bacterial “diDNases” - the technical term - could play an important role in the immune defense of bacteria. The team recently published their findings in the renowned journal “Nucleic Acids Research”, and the publication was awarded the “Breakthrough Article” designation.
Both the synthesis and degradation of large molecules by enzymes are crucial for the growth and survival of all life forms. A team led by Holger Sondermann from the Center for Structural Systems Biology (CSSB) at DESY has now discovered a new class of enzymes in bacteria and published important findings on the biochemical and structural mechanisms of these diDnases. Bioinformatic studies also suggest that these enzymes could play an important role in the immune defense of bacteria against hostile viruses.
What distinguishes diDNases from other enzymes is their highly specific mode of action. DiDNases are specialised molecular scissors and only cut small pieces of DNA and ignore RNA, a molecule that is structurally very similar to DNA. This specificity is due to the unique architecture of the enzymes' active site. “While RNA dinucleotides are well-established as signaling molecules in many biological systems, this study positions DNA dinucleotides as previously unrecognised players in cellular processes,” explains Sofia Mortensen, postdoctoral scientist in Sondermann's group and lead author of the study.
Published in the journal 'Nucleic Acids Research' as a “Breakthrough Article,” this study reveals key insights into the biochemical and structural mechanisms underlying the specificity of diDNases. Unlike related enzymes that are known to degrade both RNA and DNA, diDNases focus exclusively on DNA dinucleotides. The structural analysis, based on data collected at beamline P11 of PETRA III, highlights unique features in the enzymes’ active sites that enable them to recognise DNA while excluding RNA substrates.
The researchers at DESY were able to carry out the detailed analysis at beamline P11 which is designed for the investigation of biomolecular structures: X-ray crystallography. This involves growing crystals of the proteins and enzymes under investigation and then examining them with X-rays. “In order to uncover the molecular basis of enzyme specificity, we needed high-resolution crystal structures,” says Sondermann, last author of the study and lead scientist at DESY. “Our findings not only reveal a novel enzyme family but also imply a biological role of DNA-based dinucleotides for cellular functions.”
The discovery of diDNases provides new insights into the diversity and adaptability of bacterial systems. “We have discovered that the genes encoding diDNases are predominantly located in mobile genetic elements, genetic material that can move around and be transferred from organism to organism,” says Sondermann. “The specific location of the diDNase genes suggest a role of these enzymes in bacterial immunity, potentially protecting cells against invading viruses by targeting specific DNA substrates.”
The research team is already planning further studies to better understand the role of the newly discovered enzyme class.
Further Information:
This study is based on a close collaboration involving the groups of Drs. Vincent T. Lee and Wade C. Winkler at the University of Maryland, USA. The research was supported by grants from the U.S. National Institutes of Health and the European Union’s Horizon 2020 programme. Nucleic Acids Research (NAR), published by Oxford University Press, selected this work as a Breakthrough Article for its exceptional contribution to the fields of biology and biochemistry. Breakthrough Articles represent the highest-caliber research, addressing long-standing challenges or offering groundbreaking insights that pave the way for future discoveries.
(from DESY News)
Reference:
Mortensen S, Kuncová S, Lormand JD, Myers TM, Kim S-K, Lee VT, Winkler WC, Sondermann H, Structural and bioinformatics analyses identify deoxydinucleotide-specific nucleases and their association with genomic islands in gram-positive bacteria, Nucleic Acids Research, (2025) DOI: 10.1093/nar/gkae1235