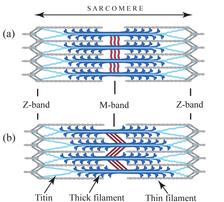
Figure 1: Diagram showing the assembly of the sarcomere. (a) Relaxed sarcomere, (b) after isometric contraction causing tension in the M-band and sliding between thick filaments (from Tskhovrebova and Trinick [7]).
- Max F. Perutz Laboratories, University of Vienna, Campus Vienna Biocenter 5, A-1030 Vienna, Austria
- European Molecular Biology Laboratory Hamburg, Hamburg, Germany,
- Section of Structural Biology, The Institute of Cancer Research, Chester Beatty Laboratories, London, United Kingdom,
- Institute of Physical Chemistry, National Centre for Scientific Research Demokritos, Athens, Greece,
- Institute for Biophysics and Munich Center for Integrated Protein Science, Physics Department, Technical University of Munich, Garching, Germany
Published as: “Superhelical Architecture of the Myosin Filament-Linking Protein Myomesin with Unusual Elastic Properties”. PLoS Biol 10(2): e1001261. doi:10.1371/journal.pbio.1001261
The process by which active muscles can generate substantial mechanical forces and still remain elastic and keep their overall organisation presents an interesting problem and one that is still not fully understood. Using a hybrid structural approach we have been able to determine the architecture of a section of the muscle filament linking protein myomesin. The data show that myomesin is folded into a superhelical coil arrangement and acts as a highly elastic ribbon that can be stretched up to 2.5 times its original length. This mechanism has never been observed before and could explain how muscles can behave elastically while still keeping their overall structural organisation.
Active muscles are capable of generating extensive mechanical forces by cycles of contraction and relaxation. These processes rely on molecular structures of extreme stability that ensure the muscle fibres resume and maintain an ordered state despite being subjected to these extreme forces. Striated muscle, found in skeletal and cardiac muscle cells, is a sophisticated and highly efficient machinery which transforms chemical energy into movement. It represents a highly organized cellular system for studying and understanding just how active forces can be generated while the overall structural organization of the underlying sarcomeric units is maintained.
The fundamental unit of the muscle cell, the sarcomere, is a highly ordered assembly consisting of filament systems (namely the thick filament composed of myosin, the thin of actin and the elastic of titin) and a number of bands or discs associated with these filaments making up the cytoskeleton. This cytoskeleton consists of three basic structural elements – alpha-actinin, which anchors the thin filaments in the so-called Z-discs at the periphery of the sarcomere, myomesin, which crosslinks the thick filaments in the central M-band, and titin. The M-band ensures the packing regularity of the thick filaments and the uniform distribution of the tension across the myosin filaments during contraction (Fig. 1).
Upon activation of the muscle cell, both substantial axial and radial forces are generated within the overall sarcomere structure [1]. To maintain a constant sarcomere volume under defined physiological conditions, these forces can lead to changes in both radial and longitudinal contour dimensions of the sarcomere. Under typical tension conditions, M-band-associated thick filaments can substantially move away from the sarcomeric center by 0.1 μm or more, which can lead to M-band-induced instability of the sarcomere [2]. Due to the presence of a stiff Z-disk architecture at the sarcomeric periphery, the amount of movement decreases with the overall sarcomere length so that the resting tension stays constant. In cardiac muscles, elastic M-band motions are thought to correlate with heart beat rate [3], rendering investigations of the underlying molecular parameters highly relevant to heart and skeletal muscle research.
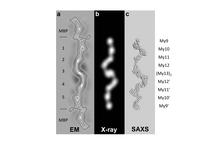
To ensure that the sarcomere retains its shape and form, there are two principal structural compartments with elastic properties. One of these two sites for molecular elasticity is within the M-band, in which so-called M-bridges transversely connect thick filaments with each other and with titin filaments [4]. At the molecular level, M-bridges are thought to be primarily composed of myomesin (MYOM1), which is universally expressed, and two related isoforms, MYOM2 and MYOM3, which show tissue-specific expression [5]. However, in the absence of ultrastructural data at a molecular resolution, the overall organisation of M-bridges, and any associated requirements for molecular elasticity, has remained largely unknown to date. To address this we have made use of a previous prediction suggesting that the entire C-terminal part of the myomesin filament consists of an array of repetitive Ig domains followed by exposed α-helical linkers [6]. By using a number of structural biology methods such as X-ray crystallography, electron microscopy and small angle X-ray scattering, we were able to determine the architecture of the C-terminal immunoglobulin domain array of myomesin (Fig. 2). The data reveal a dimeric tail-to-tail filament structure of about 360 Å in length which is folded into an irregular superhelical coil arrangement of virtually identical α-helix/domain modules (Fig. 2,3). Additional atomic force microscopy studies show that this part of myomesin can be stretched to ca. 2.5 times its original length by reversible unfolding of the α-helical linkers (Fig. 3). The findings demonstrate that the protein can adapt its overall length to dimensions that match those observed in working myofibrils. The data also demonstrate how repetitive domain modules may generate highly elastic protein structures, and future studies will determine whether this mechanism is unique to myomesin or whether it also occurs in other filament systems.
| References | ||||||||||||||
|
| Contact information |
|
Matthias Wilmanns |
| Further Information |