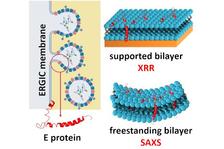
XRR and SAXS data show that the SARS-CoV-2 envelope protein condenses and bends the ERGIC membrane. (From the original publication licensed under the Creative Commons Attribution License 4.0 (CC BY))
The COVID-19 pandemic has a severe ongoing impact on society. Therefore, the development of efficient treatment is crucial for the future while our understanding of the life cycle of the SARS-CoV-2 virus and its interaction with its cell host is still comparatively limited. Already since the very beginning of the pandemic, an international team of scientists from DESY, Leipzig University (Germany), University of Vienna (Austria) and Nanyang Technological University (Singapore) has focused on the encapsulation process of the SARS-CoV-2 virus inside the infected cell and has now uncovered the physical mechanism behind this process. This was made possible by fast-track access to beamtime at DESY’s light sources to advance SARS-CoV-2 research as soon as possible after the outbreak of the pandemic. Their findings have been published in the journals Journal of Applied Crystallography and Langmuir.
A coronavirus consists of an 80 nm RNA-based viral core and a lipid bilayer-based envelope in which its spike (S), envelope (E) and matrix (M) proteins are embedded. The encapsulation of the core by the envelope occurs at the ‘endoplasmic reticulum Golgi intermediate compartment’ (ERGIC) of the infected cell. The E protein, an α-helical transmembrane protein of 76 amino acids, is pivotal for this process. Only in its presence the viral core can ‘rob’ the ERGIC membrane as its own jacket followed by the virion budding.
The impact of the E protein on the structure of the ERGIC membrane was unveiled by a combination of X-ray reflectometry (XRR), small angle X-ray scattering (SAXS) and grazing incidence X-ray off-specular scattering (GIXOS) at the PETRA III beamlines P08, P12 (operated by EMBL) and P23. “From the beginning, we were suspecting that the E protein must be able to alter the ERGIC membrane structure and its stiffness since the encapsulation of the 80 nm core would require a strong membrane curvature and hence would be energetically unfavourable” says Chen Shen, the beamline scientist at P08.
The results show that the transmembrane E protein largely condenses the lateral packing of the ERGIC membrane and hence stiffens the membrane. Moreover, the membrane is spontaneously curved in the presence of the E protein. The membrane curvature and condensation caused by the E protein should largely reduce the energy barrier of the encapsulation. “We were excited that we could get very detailed structural information to understand the role of the envelope protein”, says Chen. “We had to develop a new approach to obtain the electron density profile of the lipid bilayers from SAXS data at the required accuracy using the input from GIXOS on lipid monolayers. This helped us to get the area per lipid in the membrane, an important parameter for understanding the functions of biomembranes. This method could be very useful for other groups that study biological membranes”
Viruses with a lipid bilayer-based envelope include coronaviruses but also for instance Influenza A virus, HIV, ebolavirus. Their envelope proteins share a similar structure. The findings of this international collaboration will contribute to a deeper understanding of the role of the viral E protein in general, and hence this collaboration will be continued.
References:
Wölk, C.; Shen, C.; Hause, G.; Surya, W.; Torres, J.; Harvey, R. D.; Bello, G. Membrane Condensation and Curvature Induced by SARS-CoV-2 Envelope Protein. Langmuir 2024, 40 (5), 2646-2655. DOI: 10.1021/acs.langmuir.3c03079.
Harvey, R. D.; Bello, G.; Kikhney, A. G.; Torres, J.; Surya, W.; Wölk, C.; Shen, C. Absolute scattering length density profile of liposome bilayers obtained by SAXS combined with GIXOS: a tool to determine model biomembrane structure. J Appl Crystallogr 2023, 56 (6). DOI: 10.1107/S1600576723008439.