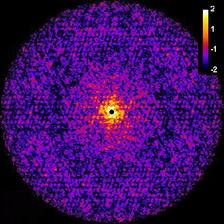
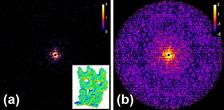
Central cut through 3D intensity distribution (see fig.2b). (Credit: Authors)
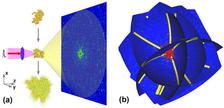
Single-particle coherent diffraction imaging is one of the promising new techniques for the investigation of biological samples to sub-nanometer resolution. It has become possible only recently due to the development of X-ray free-electron lasers (XFELs), which produce ultra-short (10–100 fs), coherent X-ray pulses with high intensity. More than 1012 photons are produced in a single FEL pulse. Very short and intense pulses are required to avoid a radiation damage of biological particles during the pulse propagation and to produce a strong scattered signal. However, after the pulse propagation the particles explode, and only one projection of the sample can be measured. This problem can be overcome by injecting particles one after another with random orientations and collecting a set of diffraction patterns. One of the challenges in processing the data from such an experiment is determining the correct orientation of each diffraction pattern from samples randomly injected in the FEL beam. A method to determine the orientation of the particle, corresponding to each diffraction pattern, is the main subject of this work .
The authors from DESY propose a method for the angular orientation determination in single-particle coherent imaging experiments based on the common arc algorithm. They obtained a significant improvement of this approach by introducing a simultaneous analysis of the common arcs for several diffraction patterns. This gives the possibility of applying the method to data with a low signal as well as to skip the classification step which is required for other orientation determination methods. The efficiency of the algorithms is demonstrated for two biological samples, an artificial protein structure without any symmetry and a virus with icosahedral symmetry.
This new method of analysing data from single-particle diffraction imaging experiments has advantages compared to other methods and may help to reach sub-nanometre resolution in 3D imaging of biological specimens in the future.
(from authors and IOP Science, the material is covered by a CCBY License)
Original Paper: O M Yefanov and I A Vartanyants, Orientation determination in single-particle x-ray coherent diffraction imaging experiments, J. Phys. B: At. Mol. Opt. Phys. 46 164013 (2013). DOI:10.1088/0953-4075/46/16/164013 (special issue of Journal of Physics B: Atomic, Molecular and Optical Physics dedicated to the frontiers of free-electron laser science).