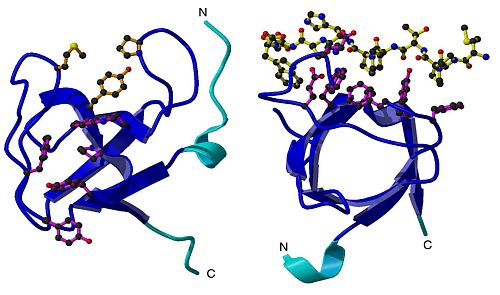
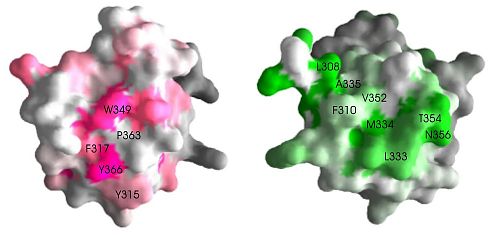
Fig. 1.
Alice Douangamath1, Fabian V. Filipp2, Andr? T.J. Klein3, Phil Barnett3, Peijian Zou1, Tineke Voorn-Brouwer3, M. Cristina Vega1, Olga M. Mayans1, Michael Sattler2, Ben Distel3 and Matthias Wilmanns1
1EMBL-Hamburg, c/o DESY, Notkestrasse 85, D-22603 Hamburg, Germany.
2EMBL-Heidelberg, Structural and Computational Biology Programme, Meyerhofstrasse 1, D-69117 Heidelberg, Germany.
3Academic Medical Center, University of Amsterdam, Department of Biochemistry P.O. Box 22700, 1100 DE Amsterdam, The Netherlands.
Published in: Mol. Cell 10, 1007-1017, 2002
While the function of most small signalling domains is confined to binary ligand interactions, the peroxisomal Pex13p SH3 domain has the unique capacity of binding to two different ligands, Pex5p and Pex14p. We have used this domain as a model to decipher its structurally-independent ligand binding sites. By the combined use of X-ray crystallography, NMR spectroscopy and circular dichroism, we show that the two ligands bind in unrelated conformations to patches located at opposite surfaces of this SH3 domain. Mutations in the Pex13p SH3 domain that abolish interactions within the Pex13p-Pex5p interface specifically impair PTS1-dependent protein import into yeast peroxisome
Background
In order for living cells to accomplish biological functions, such as reproduction and differentiation, they perform specific processes, one of which is called a protein-to-protein interaction. For each protein, these interactions are mediated by a limited number of binding sites, called binding domains [1]. One of the most studied types is the Src homology 3 (SH3) domain [2, 3, 4].
SH3 domains
SH3 domains are involved in several cell signalling pathways that regulate cell proliferation and differentiation, making them targets for drug intervention in a number of diseases, such as different types of cancer. The main strategy for drug development is the study of the SH3 domain PXXP binding site, which is recognized as having the capacity of binding ligands in two opposite orientations [5]. Most binding domains have the capacity of binding only one ligand. The discovery of a second binding site on the same sequence can provide new opportunities to target SH3 domains with multiple-site ligands [6].
PPII
The SH3 domain holds a site that is located between two specific points: the RT and n-Src loops. This section of the SH3 domain generally binds a protein sequence that begins and ends with proline, one of the amino acids that form the subunits of proteins. This sequence is called the conserved PXXP motif, and when it binds to the site in question, it adopts a poly-proline type II (PPII) conformation [7].
Pex13p interaction with 2 ligands
An example of a protein that contains an SH3 domain is the Pex13p protein found in the peroxisome. The peroxisome is a specialized intracellular structure that plays a role in fatty acid degradation. The Pex13p protein holds an SH3 domain that has the unique capacity of binding to two different ligands: Pex5p and Pex14p.They are both part of a multi-component peroxisomal targeting signal (PTS)1-receptor complex, a membrane-bound protein that imports other proteins into the peroxisome [8, 9, 10, 11].
When studying this protein-protein interaction more carefully, scientists found that binding of the Pex13p SH3 domain to Pex14p involved the classical PPII conformation/interaction. However, in the second ligand, Pex5p, this same motif was absent, giving rise to the question that there must be a novel interaction at the Pex13p SH3 domain that has not been characterized yet. As such, when Pex13p, Pex14p, and Pex5p bind together, they form a ternary complex (three-part structure), which could be tightened by the interaction of the Pex14p and Pex5p ligands [12].
Purpose of the study
The creation of specific mutations (changes in the amino acid sequence) in the Pex13p SH3 domain attempted to demonstrate that the binding of two different ligands contribute to peroxisomal matrix protein targeting. These mutations inhibit interactions within the Pex13p-Pex5p interface, which results in the ultimate impairment of protein import through the PTS1 complex of yeast peroxisomes.
Methods
With the combined use of imaging techniques such as X-ray crystallography using synchrotron radiation, NMR spectroscopy, and circular dichroism, scientists investigated the molecular nature of the interaction. The synchrotron experiments were carried out at beam line facilities at the ESRF, Grenoble, France, and at EMBL/DESY, Hamburg, Germany.
Findings
SH3 Domain Structure
Results of the imaging studies revealed that the Pex13p SH3 domain was made up of 86 residues (protein building blocks, also known as amino acids), and that it contained five-stranded b-barrel fold common to all SH3 domains. A key structure included two insertions within the RT and n-Src loop, suggesting that these insertions may play a role in separating the binding sites for the two ligands. (Figure 1)
Pex5p and Pex14p Binding Site Characteristics
Pex5p and Pex14p bind on different sites on the SH3 domain. A surface representation of the SH3 domain shows that the two ligand binding sites are at opposite faces, suggesting independent binding. (Figure 2).
In addition, Pex5p and Pex14p bind in unrelated conformations, meaning that when they bind to SH3 domain, they adopt a motif or shape that is different from the other. For example, the binding site of Pex14p takes on the PPII conformation when bound to SH3 domain. In contrast, Pex5p binding site adopts an alpha-helical conformation (slinky/coil shape) when bound to this SH3 domain.
Pex5p and Pex14p Interaction
In addition to binding to SH3 domain, these two ligands also bind to each other once they are bound to the SH3 domain. This interaction is essential to accomplish the function of protein import from the cellular cytoplasm into the peroxisome via the PTS1 complex [8, 9, 13]. A side experiment using yeast cells showed that cells with a defective PTS1-dependent protein import show reduced growth on laboratory plates [7, 8, 9, 14, 15]. Therefore this import complex is vital to cell survival. Any disturbance in this system can reduce growth rates.
Future Considerations
While the Pex13p PXXP binding site is similar across the board among known SH3 domains, suggesting that ligand binding of PXXP sequences is a common property of these domains, the Pex5p binding site does not hold any of these conserved residues. Therefore, Pex5p binding to SH3 domain is likely to be specific to the Pex13p SH3 domain.
The alpha-helical interaction site on Pex5p is the first distinct binding site identified in the SH3 domain that is structurally separate from the conventional PXXP binding site.
The Pex13p SH3 domain may serve as a model to reveal the potential of SH3 domains to be involved in multiple unrelated interactions with ligands. Recent studies investigating protein-protein interactions are suggesting that multiple ligand interactions are a greater phenomenon than originally anticipated [4, 16, 17]. Further analyses are warranted to determine whether these interactions are competitive or independent.
In summary, the concept of multiple and independent ligand binding introduces a new dimension in the study of these domains in biological processes. From the pharmacological perspective, these findings can serve as the platform from which new agents can be developed to target SH3 domains since they are such central points to many intracellular pathways involved in cell proliferation and differentiation. The combined use of different and complementary structural biology technologies have emerged as excellent tools to determine its underlying molecular principles at atomic resolution.
| References |
|
[1] T. Pawson, J. D. Scott. Signaling through scaffold, anchoring, and adaptor proteins. Science278, 2075-2080 (1997) |
| Contact information |
|
M. Wilmanns (EMBL Hamburg) |
| Further Information |